The xarray.DataArray stucture for uvecs¶
xarrayuvecs propose a accessor for xarray.DataArray to work on “Unit VECctor that are Symmetrical” (UVECS).
In this case symetrical mean that a vector x has the same meaning than -x. Therefore is can be usefull to describe non directional orientation such as crystallographic orientation dor instance.
In this notebook we will show what can be done using xarrayuvec accessor.
import xarrayaita.loadData_aita as lda #here are some function to build xarrayaita structure
import xarrayaita.aita as xa
import xarrayuvecs.uvecs as xu
import xarrayuvecs.lut2d as lut2d
import numpy as np
import matplotlib.pyplot as plt
import matplotlib.cm as cm
import xarray as xr
Load data
# path to data and microstructure
path_data='orientation_test.dat'
path_micro='micro_test.bmp'
data=lda.aita5col(path_data,path_micro)
data
<xarray.Dataset>
Dimensions: (uvecs: 2, x: 1000, y: 2500)
Coordinates:
* x (x) float64 0.0 0.02 0.04 0.06 0.08 ... 19.92 19.94 19.96 19.98
* y (y) float64 49.98 49.96 49.94 49.92 49.9 ... 0.06 0.04 0.02 0.0
Dimensions without coordinates: uvecs
Data variables:
orientation (y, x, uvecs) float64 2.395 0.6451 5.377 ... 0.6098 0.6473
quality (y, x) int64 0 90 92 93 92 92 94 94 ... 96 96 96 96 96 97 97 96
micro (y, x) float64 0.0 0.0 0.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0 0.0
grainId (y, x) int64 1 1 1 1 1 1 1 1 1 1 1 1 ... 1 1 1 1 1 1 1 1 1 1 1
Attributes:
date: Thursday, 19 Nov 2015, 11:24 am
unit: millimeters
step_size: 0.02
path_dat: orientation_test.dat- uvecs: 2
- x: 1000
- y: 2500
- x(x)float640.0 0.02 0.04 ... 19.94 19.96 19.98
array([ 0. , 0.02, 0.04, ..., 19.94, 19.96, 19.98])
- y(y)float6449.98 49.96 49.94 ... 0.04 0.02 0.0
array([4.998e+01, 4.996e+01, 4.994e+01, ..., 4.000e-02, 2.000e-02, 0.000e+00])
- orientation(y, x, uvecs)float642.395 0.6451 ... 0.6098 0.6473
array([[[2.39476627, 0.64507369], [5.37718489, 1.04999008], [5.38905313, 1.05627326], ..., [2.05826679, 0.49654617], [5.65731024, 0.94160513], [5.68523551, 1.03218772]], [[5.35955707, 1.15837502], [5.35885894, 1.12975162], [5.3613024 , 1.11701072], ..., [2.07659274, 0.52063172], [5.64753639, 0.94073247], [5.67912685, 1.04143796]], [[5.36304773, 1.23045712], [5.36165146, 1.23132979], [2.38656322, 0.57246799], ..., ... ..., [0.62378067, 0.625526 ], [0.61784656, 0.61994095], [0.61645029, 0.61400683]], [[0.68085294, 1.12399204], [0.68050388, 1.12730816], [0.67840948, 1.13900187], ..., [0.64123397, 0.62954026], [0.62430427, 0.63529985], [0.61575216, 0.62203535]], [[0.67701322, 1.13236962], [0.67753682, 1.13690747], [0.67840948, 1.13149695], ..., [0.63652158, 0.61418136], [0.61645029, 0.63948864], [0.60981804, 0.64734262]]]) - quality(y, x)int640 90 92 93 92 92 ... 96 96 97 97 96
array([[ 0, 90, 92, ..., 0, 85, 90], [81, 82, 83, ..., 0, 84, 89], [81, 80, 3, ..., 0, 79, 88], ..., [90, 91, 91, ..., 95, 95, 95], [91, 92, 91, ..., 95, 95, 95], [92, 92, 91, ..., 97, 97, 96]]) - micro(y, x)float640.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
array([[0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], ..., [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.]]) - grainId(y, x)int641 1 1 1 1 1 1 1 ... 1 1 1 1 1 1 1 1
array([[1, 1, 1, ..., 1, 1, 1], [1, 1, 1, ..., 1, 1, 1], [1, 1, 1, ..., 1, 1, 1], ..., [1, 1, 1, ..., 1, 1, 1], [1, 1, 1, ..., 1, 1, 1], [1, 1, 1, ..., 1, 1, 1]])
- date :
- Thursday, 19 Nov 2015, 11:24 am
- unit :
- millimeters
- step_size :
- 0.02
- path_dat :
- orientation_test.dat
What is an xarrayuvecs DataArray ?¶
data.orientation.head(2)
<xarray.DataArray 'orientation' (y: 2, x: 2, uvecs: 2)>
array([[[2.39476627, 0.64507369],
[5.37718489, 1.04999008]],
[[5.35955707, 1.15837502],
[5.35885894, 1.12975162]]])
Coordinates:
* x (x) float64 0.0 0.02
* y (y) float64 49.98 49.96
Dimensions without coordinates: uvecs- y: 2
- x: 2
- uvecs: 2
- 2.395 0.6451 5.377 1.05 5.36 1.158 5.359 1.13
array([[[2.39476627, 0.64507369], [5.37718489, 1.04999008]], [[5.35955707, 1.15837502], [5.35885894, 1.12975162]]]) - x(x)float640.0 0.02
array([0. , 0.02])
- y(y)float6449.98 49.96
array([49.98, 49.96])
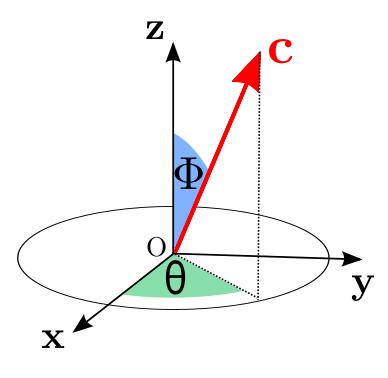
xarrayuvecs accessor should be used on xarray.DataArray da that have a dimension of \(m\times n \times 2\). The da[:,:,0] correspond to the azimuth \(\Phi\) coordinate of the unit vector and da[:,:,1] to the colatitude \(\theta\).
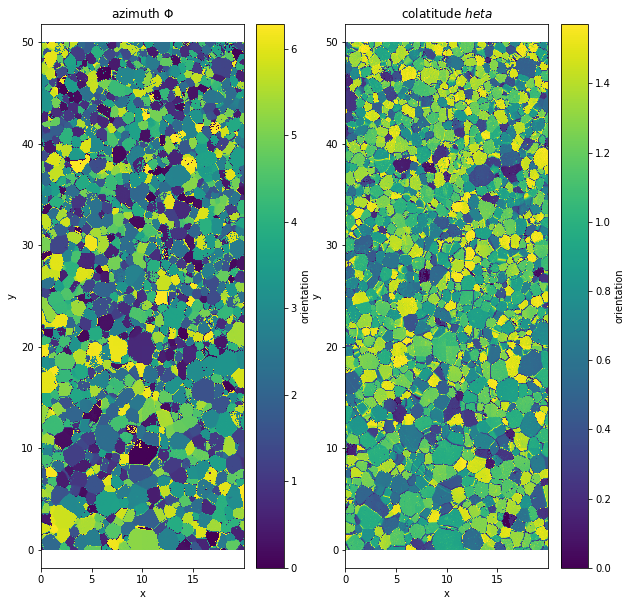
plt.figure(figsize=(10,10))
plt.subplot(1,2,1)
data.orientation[:,:,0].plot()
plt.axis('equal')
plt.title('azimuth $\Phi$')
plt.subplot(1,2,2)
data.orientation[:,:,1].plot()
plt.axis('equal')
plt.title('colatitude $\theta$')
Text(0.5, 1.0, 'colatitude $\theta$')
Export different coordinate system¶
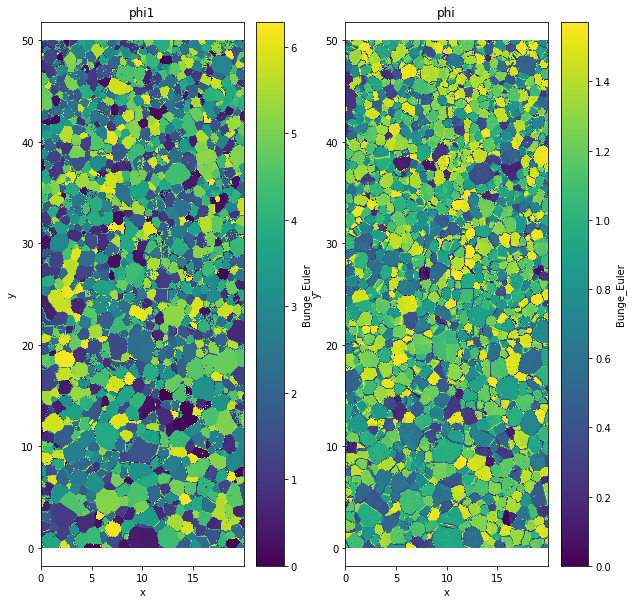
Bunge Euler convention¶
The Bunge Euler convention is widely use in crystallographic community.
\(\phi_1\) is the rotation around \(Oz\)
\(\phi\) is the rotation around \(Ou\)
data['Bunge_Euler']=data.orientation.uvecs.bunge_euler()
plt.figure(figsize=(10,10))
plt.subplot(1,2,1)
data.Bunge_Euler[:,:,0].plot()
plt.axis('equal')
plt.title('phi1')
plt.subplot(1,2,2)
data.Bunge_Euler[:,:,1].plot()
plt.axis('equal')
plt.title('phi')
Text(0.5, 1.0, 'phi')
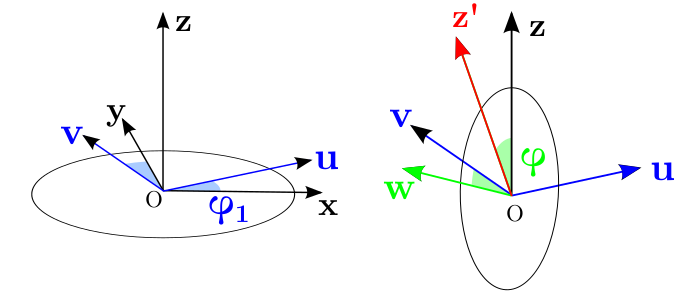
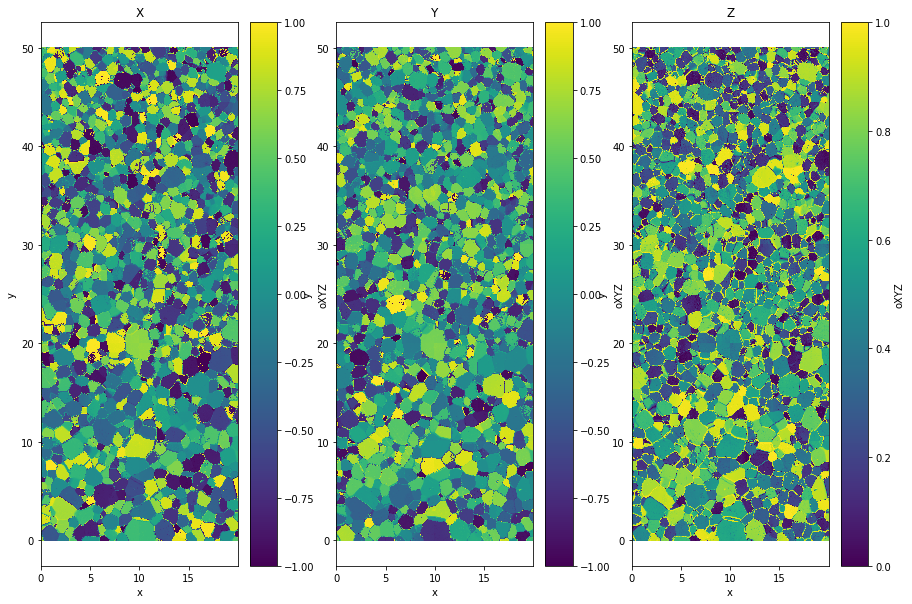
Cartesien coordinate¶
You can also extract the vector in cartesien coordinate.
data['oXYZ']=data.orientation.uvecs.xyz()
plt.figure(figsize=(15,10))
plt.subplot(1,3,1)
data.oXYZ[:,:,0].plot(cmap=cm.viridis)
plt.axis('equal')
plt.title('X')
plt.subplot(1,3,2)
data.oXYZ[:,:,1].plot(cmap=cm.viridis)
plt.axis('equal')
plt.title('Y')
plt.subplot(1,3,3)
data.oXYZ[:,:,2].plot(cmap=cm.viridis)
plt.axis('equal')
plt.title('Z')
Text(0.5, 1.0, 'Z')
Colormap¶
Colormap plotting the orientation can be done using colorwheel.
Full colorwheel¶
The colorcode used to display one orientation is choosen using a given colorwheel (show below). Using the “Full Colorwheel” will display a unique color for all the same orientation.
a black pixel display an orientation along the z-axis \((0,0,1)\)
a red pixel display an orientation along the minus x-axis \((-1,0,0)\)
a blue pixel display an orientation along the \(\left(\frac{\sqrt(2)}{2},\frac{-\sqrt(2)}{2},0\right)\)
This representation is quite usefull but is not exempt of default. For instance The color codding is discontinous. That mean that vectors \(\left(1,0,+\delta_z\right)\) and \(\left(1,0,-\delta_z\right)\) show very defferent color (pale blue and red) even if the angle between both decrease to 0 as \(\delta_z\) decrease to 0.
lut_f=lut2d.lut()
plt.figure(figsize=(3,3))
plt.imshow(lut_f)
plt.axis('equal')
plt.axis('off')
plt.title('Full colorwheel')
Text(0.5, 1.0, 'Full colorwheel')

The colormap associated with this colorwheel can be compute using xarrayuvecs
data['FullColormap']=data.orientation.uvecs.calc_colormap()
plt.figure(figsize=(5,10))
data.FullColormap.plot.imshow()
plt.axis('equal')
(-0.01, 19.990000000000002, -0.010000000000001563, 49.99000000000001)
Semi colorwheel¶
The semi colorwheel is an over represntation of the orientation. It has the advantage to not have any discountinuities but one color does not correspond to a unique orientation.
Warning
\((x,y,z)\) and \((-x,-y,z)\) share the same colorcoding.
lut_s=lut2d.lut(semi=True)
plt.figure(figsize=(3,3))
plt.imshow(lut_s)
plt.axis('equal')
plt.axis('off')
plt.title('Semi colorwheel')
Text(0.5, 1.0, 'Semi colorwheel')

data['SemiColormap']=data.orientation.uvecs.calc_colormap(semi=True)
plt.figure(figsize=(5,10))
data.SemiColormap.plot.imshow()
plt.axis('equal')
(-0.01, 19.990000000000002, -0.010000000000001563, 49.99000000000001)
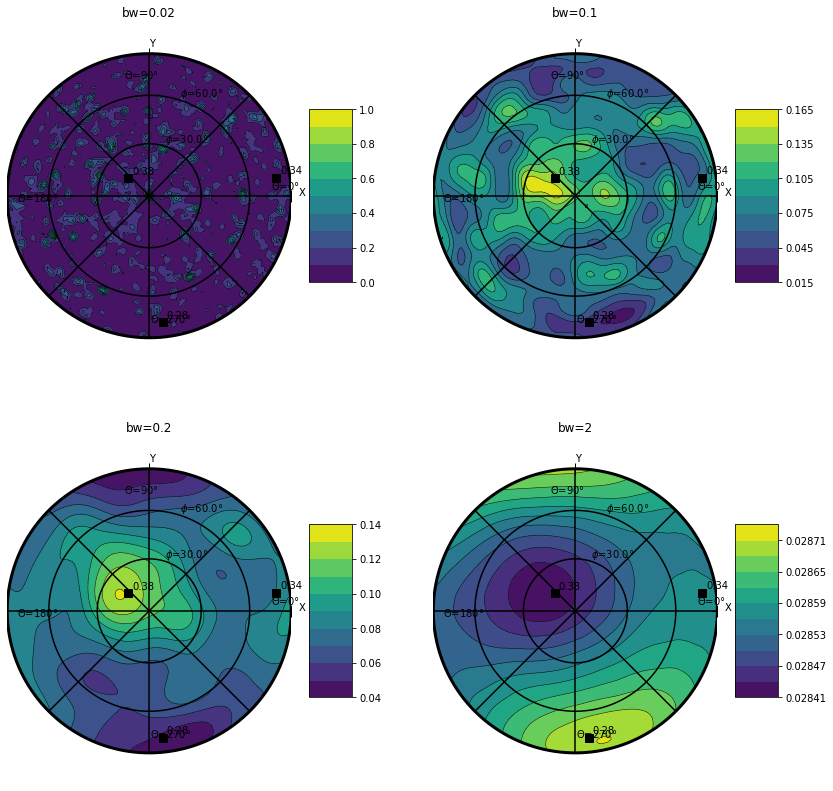
The ODF¶
The Orientation Density Function (ODF) is a probability density function for orientation. Therefore it’s integral over the sphere is equal to 1.
It is evaluated using a Kernel Density Estimation (KDE) with a gaussian kernel.
Warning
Therefore the representation is quite sensitive to the ‘bandwidth’ bw paramerter of the gaussian kernel and should be choosen carefully.
plt.figure(figsize=(14,14))
plt.subplot(221)
data.orientation.uvecs.plotODF(bw=0.02,cmap=cm.viridis)
plt.title('bw=0.02')
plt.subplot(222)
data.orientation.uvecs.plotODF(bw=0.1,cmap=cm.viridis)
plt.title('bw=0.1')
plt.subplot(223)
data.orientation.uvecs.plotODF(bw=0.2,cmap=cm.viridis)
plt.title('bw=0.2')
plt.subplot(224)
data.orientation.uvecs.plotODF(bw=2.,cmap=cm.viridis)
plt.title('bw=2')
Text(0.5, 1.0, 'bw=2')