The xarray.Dataset strucuture for AITA¶
xarrayaita is a dataset accessor for xarray. In this section we explain what is minimal contruction that you need to build and what are the function that you can applied through this xarrayaita
The minimal structure of xarrayaita¶
import xarrayaita.loadData_aita as lda #here are some function to build xarrayaita structure
import xarrayaita.aita as xa
import numpy as np
import matplotlib.pyplot as plt
import matplotlib.cm as cm
import xarray as xr
Load data to show an exemple.
# path to data and microstructure
path_data='orientation_test.dat'
path_micro='micro_test.bmp'
In this case we use a constructor function that take as input :
a
.datfile that is a output from G50 fabric analyser. If you have an over analyser it should be easy to implement an other constructor function.a
.bmpthat is the microstructure image if it as been processing elsewhere (grains boundaries are the white pixel). This input can be leave empty
help(lda.aita5col)
Help on function aita5col in module xarrayaita.loadData_aita:
aita5col(adr_data, micro_adress=0)
Function to load the data from G50 analyser that have 5 columns
:param adr_data: path to the data
:type adr_data: str
:param micro_adress: path to microstructure file (black and white image where grains boundaries are white) `.bmp`
:type micro_adress: str
data=lda.aita5col(path_data,path_micro)
xarray.Dataset¶
The data strucutre is a regular xarray.Dataset. Therefore it can be use as one.
Specific function can be applied to it :
data.xa.FUNCTION
You can visualised what is store in your xarray.Dataset using :
data
<xarray.Dataset>
Dimensions: (uvecs: 2, x: 1000, y: 2500)
Coordinates:
* x (x) float64 0.0 0.02 0.04 0.06 0.08 ... 19.92 19.94 19.96 19.98
* y (y) float64 49.98 49.96 49.94 49.92 49.9 ... 0.06 0.04 0.02 0.0
Dimensions without coordinates: uvecs
Data variables:
orientation (y, x, uvecs) float64 2.395 0.6451 5.377 ... 0.6098 0.6473
quality (y, x) int64 0 90 92 93 92 92 94 94 ... 96 96 96 96 96 97 97 96
micro (y, x) float64 0.0 0.0 0.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0 0.0
grainId (y, x) int64 1 1 1 1 1 1 1 1 1 1 1 1 ... 1 1 1 1 1 1 1 1 1 1 1
Attributes:
date: Thursday, 19 Nov 2015, 11:24 am
unit: millimeters
step_size: 0.02
path_dat: orientation_test.dat- uvecs: 2
- x: 1000
- y: 2500
- x(x)float640.0 0.02 0.04 ... 19.94 19.96 19.98
array([ 0. , 0.02, 0.04, ..., 19.94, 19.96, 19.98])
- y(y)float6449.98 49.96 49.94 ... 0.04 0.02 0.0
array([4.998e+01, 4.996e+01, 4.994e+01, ..., 4.000e-02, 2.000e-02, 0.000e+00])
- orientation(y, x, uvecs)float642.395 0.6451 ... 0.6098 0.6473
array([[[2.39476627, 0.64507369], [5.37718489, 1.04999008], [5.38905313, 1.05627326], ..., [2.05826679, 0.49654617], [5.65731024, 0.94160513], [5.68523551, 1.03218772]], [[5.35955707, 1.15837502], [5.35885894, 1.12975162], [5.3613024 , 1.11701072], ..., [2.07659274, 0.52063172], [5.64753639, 0.94073247], [5.67912685, 1.04143796]], [[5.36304773, 1.23045712], [5.36165146, 1.23132979], [2.38656322, 0.57246799], ..., ... ..., [0.62378067, 0.625526 ], [0.61784656, 0.61994095], [0.61645029, 0.61400683]], [[0.68085294, 1.12399204], [0.68050388, 1.12730816], [0.67840948, 1.13900187], ..., [0.64123397, 0.62954026], [0.62430427, 0.63529985], [0.61575216, 0.62203535]], [[0.67701322, 1.13236962], [0.67753682, 1.13690747], [0.67840948, 1.13149695], ..., [0.63652158, 0.61418136], [0.61645029, 0.63948864], [0.60981804, 0.64734262]]]) - quality(y, x)int640 90 92 93 92 92 ... 96 96 97 97 96
array([[ 0, 90, 92, ..., 0, 85, 90], [81, 82, 83, ..., 0, 84, 89], [81, 80, 3, ..., 0, 79, 88], ..., [90, 91, 91, ..., 95, 95, 95], [91, 92, 91, ..., 95, 95, 95], [92, 92, 91, ..., 97, 97, 96]]) - micro(y, x)float640.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
array([[0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], ..., [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.], [0., 0., 0., ..., 0., 0., 0.]]) - grainId(y, x)int641 1 1 1 1 1 1 1 ... 1 1 1 1 1 1 1 1
array([[1, 1, 1, ..., 1, 1, 1], [1, 1, 1, ..., 1, 1, 1], [1, 1, 1, ..., 1, 1, 1], ..., [1, 1, 1, ..., 1, 1, 1], [1, 1, 1, ..., 1, 1, 1], [1, 1, 1, ..., 1, 1, 1]])
- date :
- Thursday, 19 Nov 2015, 11:24 am
- unit :
- millimeters
- step_size :
- 0.02
- path_dat :
- orientation_test.dat
There is 4 xarray.DataArray variables :
orientation: that store the orientation of each pixel. It is store as azimuth and colatitude. (see figure below) It is meant to be use with xarrayuvecsDataArray.quality: that is the quality factor given by the analyser (between \([0,100]\)).micro: it is the microstrucure image given as input.grainId: it is a labelling of the microstrucure where each pixel within the same grain as a unique label.
This is the minimal structure to use xarrayaita function.
Visualisation of the variable¶
Therefore every Data variable can be plot using regular xarray ploting function
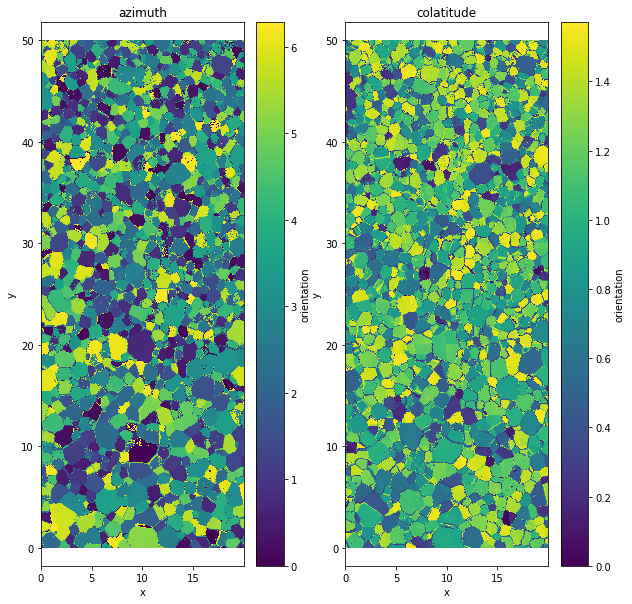
Orientation variable¶
The data are store as azimuth and colatitude
plt.figure(figsize=(10,10))
plt.subplot(1,2,1)
data.orientation[:,:,0].plot()
plt.axis('equal')
plt.title('azimuth')
plt.subplot(1,2,2)
data.orientation[:,:,1].plot()
plt.axis('equal')
plt.title('colatitude')
Text(0.5, 1.0, 'colatitude')
Quality variable¶
plt.figure(figsize=(5,10))
data.quality.plot(cmap=cm.gray)
plt.axis('equal')
plt.title('quality')
Text(0.5, 1.0, 'quality')

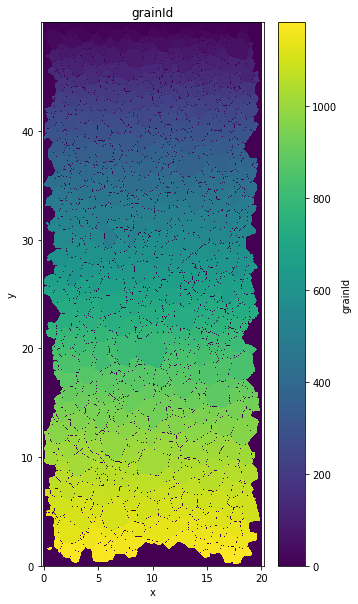
GrainID variable¶
plt.figure(figsize=(5,10))
data.grainId.plot()
plt.axis('equal')
plt.title('grainId')
Text(0.5, 1.0, 'grainId')
If you prefere to look at Bunge Euler angle you can extract them using uvecs
All the variable are visible in your xarray.Dataset
The function within xarrayaita¶
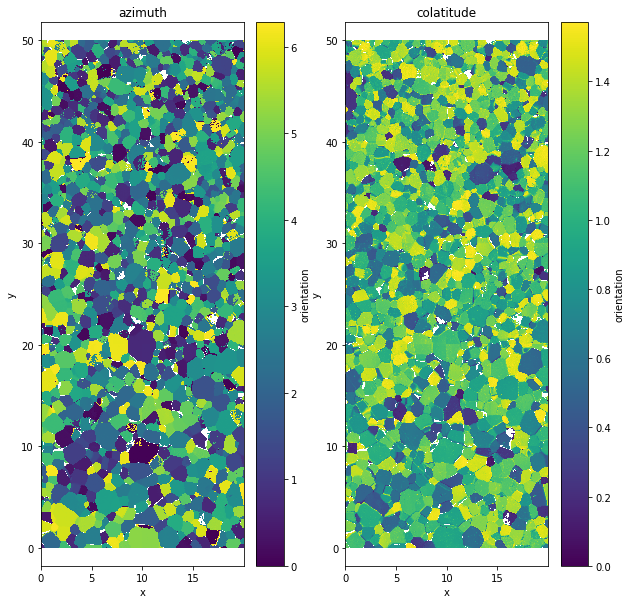
xa.filter¶
It only affect the orientation value. Where the data.quality variable is bellow the given value val the orienation value is replace by a nan.
help(xa.aita.filter)
Help on function filter in module xarrayaita.aita:
filter(self, val)
Put nan value in orientation file
data.aita.filter(75)
plt.figure(figsize=(10,10))
plt.subplot(1,2,1)
data.orientation[:,:,0].plot()
plt.axis('equal')
plt.title('azimuth')
plt.subplot(1,2,2)
data.orientation[:,:,1].plot()
plt.axis('equal')
plt.title('colatitude')
Text(0.5, 1.0, 'colatitude')
Symmetry transformation¶
fliplr¶
Flip the data with a horizontal mirror.
help(xa.aita.fliplr)
Help on function fliplr in module xarrayaita.aita:
fliplr(self)
flip left right the data and rotate the orientation
May be it is more a routation around the 0y axis of 180 degree
rot180¶
help(xa.aita.rot180)
Help on function rot180 in module xarrayaita.aita:
rot180(self)
rotate 180 degre around Oz the data and rotate the orientation
